Content
- What is FOAF and how can I use it ?
- What is SciFOAF ?
- What is MyFOAFExplorer ?
- Download
- Building
- Using SciFOAF and MyFOAFExplorer
- References
- Links
What is FOAF and how can I use it ?
The FOAF project defines a semantic format based on RDF/XML to define persons or groups, their relationships, as well as their basic properties such as name, e-mail address, subjects of interest, publication, and so on...
What is SciFOAF ?
SciFOAF is an interface used to generate a FOAF file using NCBI Pubmed. The PubMed bibliographic database is a powerful source to discover collaboration networks as each paper submitted contains potential information about each author. An interface called SciFOAF (Scientific FOAF) was written to generate a FOAF file from the literature found in PubMed. It is initialized with a PubMed identifier (PMID) that is used as a seed to start browsing a collaboration network and the user is asked to select the authors of this paper that will appear in the FOAF file. Authors are stored in a 'stack' and the user interaction will be required until this stack is empty. For each person in the stack complementary information (such as e-mail, web site...) can be fulfilled and his latest publications are fetched from NCBI using the NCBI E-Utilities tool. Then, for each article the user is asked if the current person is unambiguously one of the authors and whether he knows any of the co-authors (which may not be necessarily the case when there is a large number of collaborators) and each of these new co-author is added on top of the stack. Laboratories addresses are extracted from the articles and the user can select among this list to affiliate the current author to one or more laboratory. The user can also link the current author with a person already depicted if they know each other but did not take part in a common publication. Author’s interests are defined using the MESH terms of their papers. Once all information about the current author have been fulfilled he is removed from the stack and at the end, the user can save its FOAF file as a XML/RDF file. This process could have been generated automatically by a recursive program without human intervention if there was not ambiguity between authors names and if the addresses of the laboratories were always written the same way but this is unfortunately not the case: the same address of a laboratory can be written differently from one paper to another and there is no unique identifier that could be used to discriminate two homonymous authors within the XML description of papers. Moreover, authors’ name definition may vary from one journal to another as some journal may use the initial of an author while another may use the complete first name. Furthermore the NCBI E-Search utility just uses the family name and the initial so homonymous authors may be fetched with the same query. In consequence SciFOAF requests the user to confirm/infirm the relationships between authors and their publications.
How to use SciFOAF ?
Start your network description by a NCBI Pubmed ID (PMID). e.g. 15047801.

Select people you know and you want to include in your network
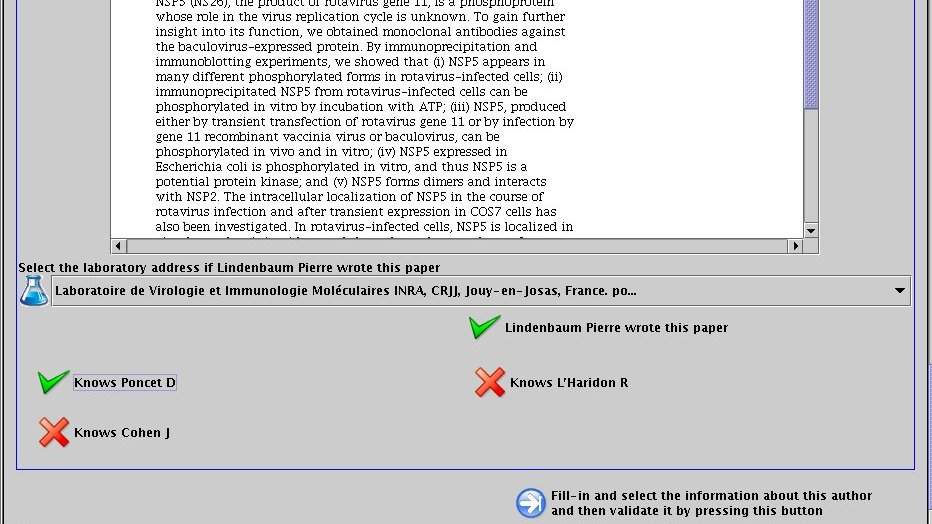
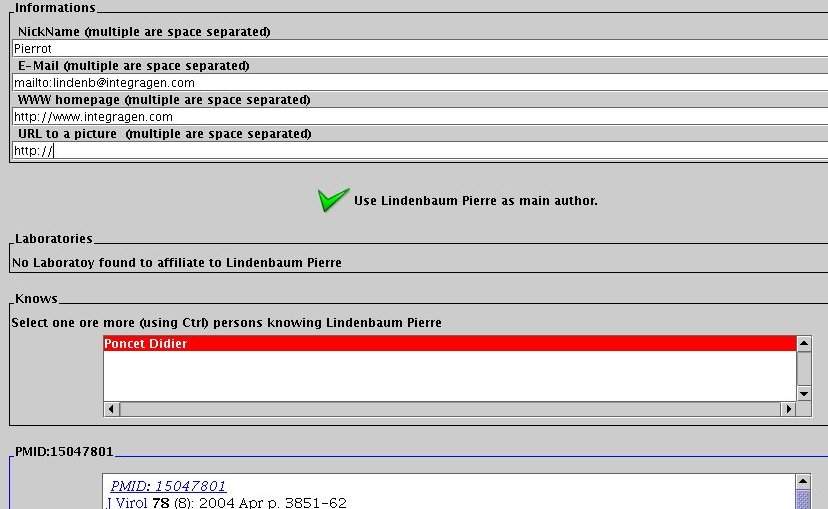
For each selected author, complete its personal information (mail, web page...) and select his affiliated laboratories
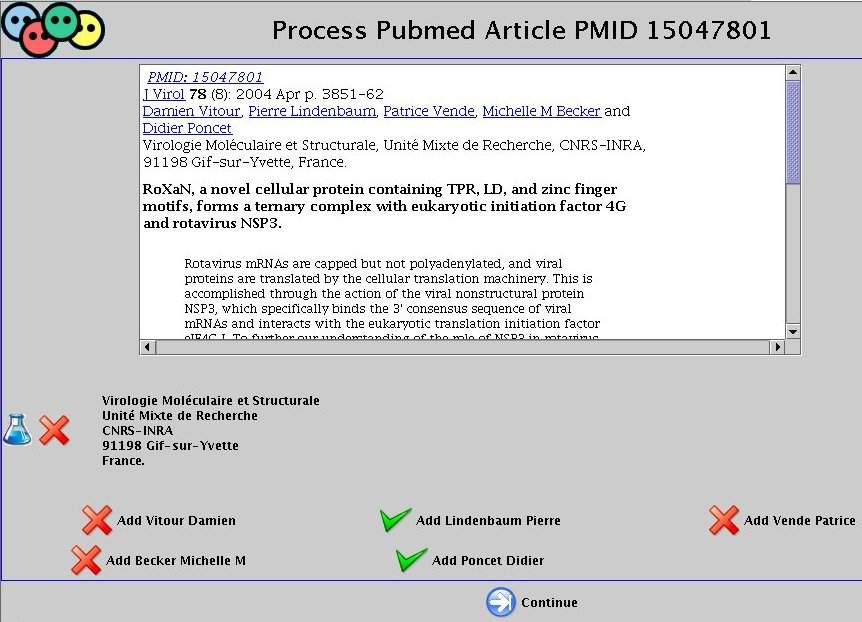
For each paper, check if the paper was written by the current author. When you're done with this author click on the 'NEXT' button at the bottom of the scrollable page
When your done (there is no more author on the stack), click on the 'Save ' button to export the RDF/XML describing the network
What is MyFOAFExplorer and how can I use it ?
MyFOAFExplorer was created after SciFOAF in order to browse a scientific network, its publications, its laboratories,... The
program is an applet that takes as input a FOAF file. The applet can
be
downloaded and used for your laboratoy/social network to introduce your staff, your publications,...The code does not contain a compliant RDF parser (such as JENA), but the way it parses the XML works fine for my test, so it could be used as a framework to write your own FOAF file..
Screenshots
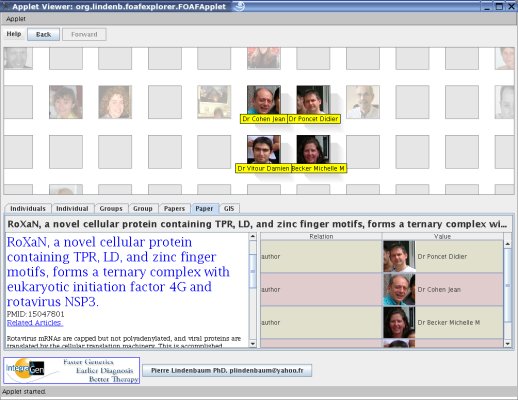
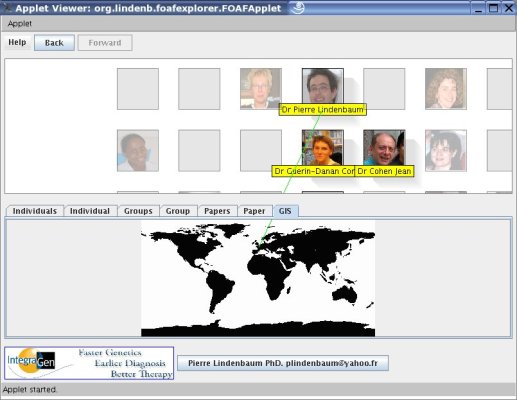
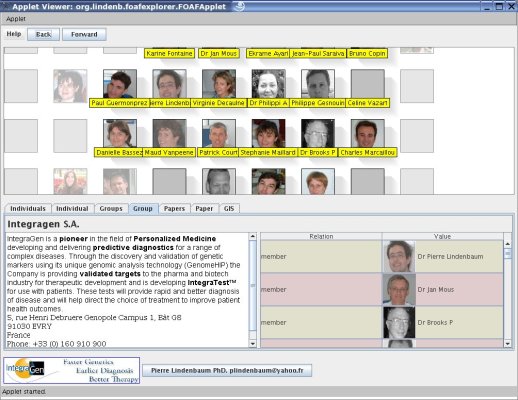
Building
ANT and JAVAC are required:
Buildfile: build.xml
compile-all:
[javac] Compiling 2 source files
jar-all:
[jar] Building jar: urbigene/urbigene.jar
all:
BUILD SUCCESSFUL
Total time: 17 seconds
Using SciFOAF and MyFOAFExplorer.
To run SciFOAF:
java -cp urbigene.jar org.lindenb.scifoaf.SciFOAF
To run MyFOAFExplorer:
java -cp urbigene.jar org.lindenb.foafexplorer.Main sample/foaf.xml
To run MyFOAFExplorer as an applet:
<applet archive="urbigene.jar" width="640" height="640" code="org.lindenb.foafexplorer.FOAFApplet"> <param name="foaf" value="foaf.xml"> </applet>
References
- Bachrach,C.A. and Charen,T. (1978) Selection of MEDLINE contents, the development of its thesaurus, and the indexing process. Medical Informatics (London), 3,237-254.
- Bohne-Lang,A., Lang,E. (2005) Do we need a Unique Scientist ID for publications in biomedicine? Biomed Digit Libr., Mar 22;2(1):1.
- Newman,M.E. (2001) The structure of scientific collaboration networks. Proc. Natl. Acad. Sci. USA., 98,404-409.
- Ding,L., Zhou L., Finin T. and Joshi A. (2005) How the Semantic Web is Being Used: An Analysis of FOAF Documents. Proceedings of the 38th International Conference on System Sciences.
- Wheeler,D.L., Barrett,T., Benson,D.A., Bryant,S.H., Canese,K., Church,D.M., DiCuccio,M., Edgar,R., Federhen,S., Helmberg,W., Kenton,D.L., Khovayko,O., Lipman,D.J., Madden,T.L., Maglott,D.R., Ostell,J., Pontius,J.U., Pruitt,K.D., Schuler,G.D., Schriml,L.M., Sequeira,E., Sherry,S.T., Sirotkin,K., Starchenko,G., Suzek,T.O., Tatusov,R., Tatusova,A., Wagner,L., Yaschenko,E. (2005). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 33, 39-45.